

Document Type : Research Article (Regular Paper)
Authors
1 Department of Animal Sciences, College of Agriculture, Isfahan University of Technology, Isfahan 84156-83111, Iran.
2 Department of Animal Science, Faculty of Agriculture, Shahid Bahonar University of Kerman, Kerman, Iran.
Abstract
Keywords
Main Subjects
Introduction
Rapid development in molecular genetics has led to detection of many polymorphic sites throughout the cattle genome in recent years, which can be used in marker-assisted selection programs (Huang et al., 2006; Pagan et al., 2006; Javanmard et al., 2008; Shojaei et al., 2011; Kharrati Koopaei et al., 2012 and Pasandideh et al., 2015). Lactoferrin (LTF) is an approximately 80-kDa iron-binding glycoprotein present in milk and such external secretions of the body, as saliva, bile, tears, sperm and vaginal fluids (Rainard 1993 and Schanbacher et al., 1997).The protein is a monomeric metal-binding glycoprotein naturally produced by secondary granules and mammary epithelial cells in response to inflammatory stimuli, such as health problems of mammary glands. The molecule is folded into two homologous lobes; N and C, each divided into two metal-binding sites (N1 and N2, C1 and C2) and binds one free iron in a deep cleft between two domains (Anderson et al., 1989; Nuijenset al. 1996; Moor et al., 1997 and Kaminski et al., 2006). In cattle, LTF gene is localized in chromosome 22 and consists of 17 exons and spreads on about 34.5 kbp of a genomic DNA (Schwerin et al., 1994). LTF gene is highly polymorphic and it has been shown that some of its variants are related to milk production traits and mastitis resistance in cattle (Kaminski et al., 2006; Wojdak-Maksymiec et al., 2006; Kaminski et al., 2008 and Pawlik et al., 2014).
The objective of this study was to investigate the association of LTF gene variants with some milk production and reproductive traits in Iranian Holstein dairy cows.
Materials and methods
Animals and data collection
The study included 408 randomly selected Iranian Holstein cows belonging to five large dairy herds in Isfahan province, Iran. All cows were kept in similar environmental conditions. The phenotypic data were collected for the following production and reproductive traits: 305-day milk yield (Kg), fat content (%), protein content (%), pregnancy length (d), milk days (d) and SCS. Three- to four-years accurate data of each cow was used for analysis of the selected traits. The analysis of interested traits was done on the basis of average values for each individual. Blood samples were collected in venoject vacuum tubes containing EDTA and stored at −20 ° C until DNA extraction.
SNP genotyping
Genomic DNA was extracted from whole blood samples using the salting out protocol (Miller et al., 1988) and LTF gene was amplified with standard PCR (Thermo cycler, BIOMETRA, Germany). The PCR mixture contained 50 ng genomic DNA, 10 pmol of each primer, 2 μL 10x PCR buffer, 1.5 mM MgCl2, 200 μmol dNTPs, and 1 unit of TaqDNA polymerase, in a total volume of 20 μL.The sequences of primers (Wojdak –Maksymiecet al., 2006) were as follows:
Forward: 5'- GCC TCA TGA CAA CTC CCA CAC -3'
Reverse: 5'- CAG GTT GAC ACA TCG GTT GAC -3'
The 301 bp fragment of the bovine LTF gene was amplified using PCR under the following conditions: initial denaturation at 94 °C for 2 min, 32 cycles of denaturation at 94 °C for 60 s, 61°C annealing temperature for 45 s, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. Digestion of PCR products of 301 bp was carried out with 5 U of EcoRI enzyme (Fermentas, St. Leon-Rot, Germany) in a reaction volume of 20 μL at 37 °C for 6 h. Digested fragments were visualized on a 2% agarose gel after electrophoresis.
Statistical analysis
Test for deviation from Hardy-Weinberg equilibrium was performed using the POPGENE software (Nei1977). The association of genotypes and traits of interest was analyzed using GLM procedure of SAS 9.1 software (SAS Institute Inc., Cary, NC, USA) for production and reproductive traits. Least squares means (LSMeans) of the genotypes were compared by the LSMeans test. The statistical models were as follows:
Yijkl1= μ + Gi+ HYSj + Sk+b1(Nijkl + N) + b2(zijkl+ Z) + eijkl
Yijkl2= μ + Gi+ HYSj + Sk+ b1 (Nijkl + N) + eijkl
Where, Yijkl1- the phenotypic value for each milk-related trait,Yijkl2– the phenotypic values of the reproductive traits, μ – overall mean, Gi – effect of genotype, HYSj– fixed effect of herd (1, 2, 3, 4, and 5), year and season of parturition, Sk– random effect of sire (1,…,155), b1– the linear regression coefficient of dry period, Nijk– effect of dry period, b2– the linear regression coefficient of open days trait, zijk– effect of open days, eijkl– random error.
Results
Allelic and genotypic frequencies
The PCR resulted in amplification of a specific fragment with the expected size of 301 bp (Figure 1). The DNA restriction fragments were obtained for LTF gene using EcoRI enzyme (Figure 2). The undigested fragments (301 bp) represented the AA genotype, the fragments of 201 and 100 bp length showed the AB genotype, and the BB genotype was not detected. Both A and B alleles were observed in all five herds with overall frequencies of 0.803 and 0.197, respectively (Table 1). In all herds, the AA genotype was the most frequent and the value of chi-square was greater than the critical value (P < 0.05), since it has been under selection for milk production traits for years.
Figure 1. Agarose gel electrophoresis of PCR products
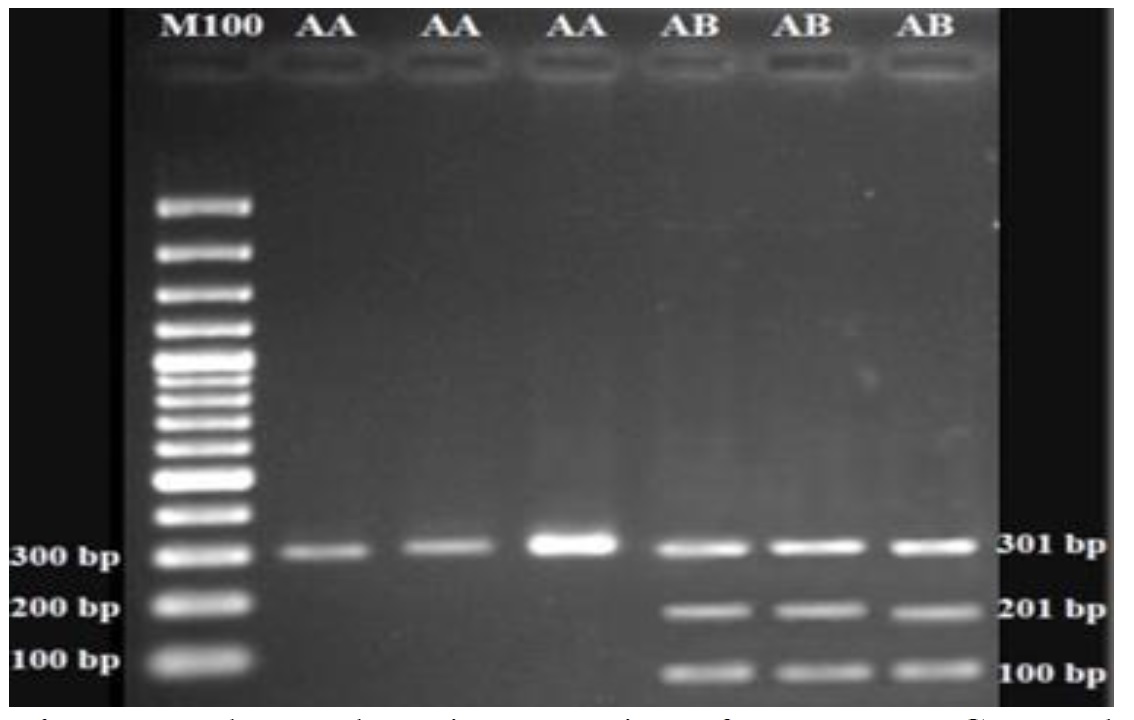
Figure 2. Electrophoretic separation of LTF gene PCR products digested with EcoR1
Table 1. Allelic and genotypic frequencies of the LTF variants in Iranian Holstein cattle population
|
Herd |
Number of animals |
Allele |
|
Genotype |
Chi-square |
||
|
A |
B |
|
AA |
AB |
|||
|
1 |
65 |
0.78 |
0.22 |
|
0.550 |
0.450 |
5.14* |
|
2 |
96 |
0.79 |
0.21 |
|
0.570 |
0.430 |
6.87** |
|
3 |
76 |
0.81 |
0.19 |
|
0.618 |
0.381 |
4.06* |
|
4 |
83 |
0.83 |
0.17 |
|
0.663 |
0.337 |
3.27* |
|
5 |
84 |
0.81 |
0.19 |
|
0.620 |
0.380 |
4.48* |
|
Total |
404 |
0.803 |
0.197 |
|
0.606 |
0.393 |
24.07** |
**P –significant at < 0.01; *P –significant at p < 0.05.
Association analysis
Table 2 shows the explored effects of LTF genotypes upon milk production and selected reproductive traits. Results showed that the SNP located in intron 6 of the LTF gene was associated with milk fat percentage and SCS (P 0.1) was evident between the LTF polymorphism and 305-day milk yield, protein percentage, pregnancy length (d), and milk days (d).
Table 2. Least square means and standard errors (LS Mean± SE) of the production and reproductive traits in two different LTF genotypes in Iranian Holstein cattle population.
|
Productive traits |
Genotype |
|
|
AA |
AB |
|
|
305-day milk yield (Kg) |
9535.92 ± 130.77 |
9379.44 ± 153.22 |
|
Fat (%) |
3.159 ± 0.04 a |
3.291 ± 0.05a |
|
Protein (%) |
2.961 ± 0.02 |
2.985 ± 0.02 |
|
Reproductive traits |
|
|
|
Pregnancy length (d) |
276.31 ± 0.44 |
276.79 ± 0.52 |
|
Milk days (d) |
315.60 ± 1.07 |
315.15 ± 1.27 |
|
SCS |
1.78 ± 0.07 b |
1.97 ± 0.09b |
Different superscript letters indicate significant differences between the genotypes (P < 0.05).
Discussion
This study reports the association of LTF gene polymorphism with some milk production and reproductive traits in Iranian Holstein cows. The LTF gene was chosen because of its expression in mammary glands, involvement in mammary health and its biological functions in the immune system response and inflammatory process (Rainard 1993; Schanbacheret al., 1997 and Chanetonet al., 2008).
The SNP in the sixth intron of bovine LTF gene was first discovered by Seyfert and Kuhn (1994). The researchers found two alleles, A and B, which encoded three possible genotypes (AA, AB and BB). In our study, two genotypes, AA and AB, of the LTF gene were observed, while BB genotype was not detected. This may be due to different genetic backgrounds of the animals in these studies. Different breeding goals and selection criteria will lead to differences in genetic background over generations.
It was shown that this SNP was associated with fat percentage and SCS in milk. Results showed that the B allele was associated with high SCS in milk, as animals carrying genotype AA were found to have significantly lower SCS in comparison with AB genotype.
A pronounced influence of LTF on SCC has been reported in some cattle populations. For example, Wojdak - Maksymiec et al. (2006) reported an association between LTF gene and SCC in Polish Black-and-White dairy cows. They claimed that AB heterozygous animals had a higher SCC than homozygous AA. In a study of the same SNP by Sender et al. (2006), BB homozygous animals had the lowest SCC and the AB heterozygotes had the highest. However, Srubarova and Dvorak (2009) reported no significant association between LTF variants and SCC in milk.
Recently, Huang et al. (2010) investigated three SNPs in the 5′- flanking region and one SNP in exon 1 of LTF gene in Chinese Holstein cattle and suggested that two major haplotypes (EFCDBB and GGEFDD) may be potential candidates for low SCS.
In the present study, the B allele was associated with higher milk fat percentage (Pet al. (2014) reported significant effects of the LTF polymorphisms on milk yield and milk composition traits in Polish Holstein cattle. They showed that 4 SNPs of the bovine LTF (LTF-926, LTF+32, LTFex4 and LTFex9) influenced fat yield, but only one (LF+32) was associated with fat percentage.
The precise molecular mechanisms by which LTF affects milk fat percentage remain to be established. However, in this research the results indicated that LTF genotypes had statistically significant effect on this trait (P < 0.05). A possible hypothesis is that this polymorphism indirectly affects milk fat percentage by being in linkage disequilibrium with another polymorphism that directly influences the studied trait. This hypothesis is supported by the fact that several quantitative trait loci (QTL) for milk fat percentage, milk protein percentage and SCS were detected close to the LTF gene on BTA 22 (Boichardet al., 2003 and Ashwellet al., 2004). According to Rodriguez-Zas et al. (2002), some markers located on BTA 22 had a significant influence on milk fat percentage. In addition, Harder et al. (2006) reported one QTL affecting persistency of milk fat and protein yield between LTF gene and HMH1R marker in German Holstein dairy cattle.
Besides, there are several SNPs in bovine LTF gene, reported to have important roles on milk yield and milk composition traits (milk protein yield, protein percentage, milk fat yield and fat percentage) in dairy cattle (Kaminski et al., 2006; Kaminski et al., 2008 and Pawliket al., 2014).
In conclusion, the present study demonstrates a significant effect of LTF gene variants on SCS and milk fat percentage. These results suggested that this SNP may be a potential genetic marker in selection programs for dairy cattle through marker-assisted selection. Further investigations are needed to confirm these results and determine the mechanisms underlying the effect of LTF gene.
Acknowledgements
The authors gratefully acknowledge Isfahan University of Technology for financial support of this research.