

Document Type : Short Communication
Authors
1 Department of Genetics and Biotechnology, Faculty of Biological Sciences,Varamin-Pishva Branch, Islamic Azad University, Varamin, PO Box 33817-74895, Iran.
2 Department of Animal Science, Faculty of Agriculture, Shahid Bahonar University of Kerman, Kerman, PO Box 76169-133, Iran.
3 Department of Biology, Faculty of Sciences, Razi University, Kermanshah, Iran.
Abstract
Keywords
Main Subjects
Introduction
Genetic polymorphism in native breeds can be useful in preservation of the genetic resources; therefore, it is important to genetically characterize the indigenous breeds (Bastos et al., 2001). Myostatin (MSTN) or growth differentiation factor-8 (GDF-8) is a member of the mammalian transforming growth factor (TGF-beta), which plays a role in the regulation of embryonic development and tissue homeostasis in adults (Sonstegard et al., 1998). It is known to block myogenesis and enhance chondrogenesis as well as epithelial cell differentiation in vitro. In mice, null mutants are significantly larger than wild-type animals, with 200-300% more skeletal-muscle mass as a result of muscle fiber hyperplasia and hypertrophy (McPherron et al., 1997). Muscular hypertrophy (mh), known as “double-muscling” in cattle, has been recognized as a physiological character for years (Arthur, 1995) and is seen in Belgian Blue and Piedmontese cattle (Kambadur et al., 1997). These animals has less bone, less fat and 20% more muscle (Hanset, 1991; Casas et al., 1998). Such a major effect of a single gene on processing yields opened a potential channel for improving processing yields of animals using knockout technology (Kocabas et al., 2002). The CAST gene is located on the chromosome 5 of sheep and plays important roles in formation of muscles and meat tenderness after slaughter. The rate and extent of skeletal muscle growth depend mainly on three factors: rate of muscle protein synthesis, rate of muscle protein degradation and the number and size of skeletal muscle cells. Calpain activity is required for myoblast fusion and cell proliferation in addition to cell growth (Barnoy et al., 1997). The calpain system may also affect the number of skeletal muscle cells in domestic animals by altering the rate of myoblast proliferation and modulating myoblast fusion. A number of studies (Forsberg et al., 1989; Goll et al., 1992; Cong et al., 1998) have shown that the calpain system is also important in normal skeletal muscle growth. Increased rate of skeletal muscle growth can result from a decreased rate of muscle protein degradation and this is associated with a decrease in activity of the calpain system, due principally to a large increase in calpastatin activity (Goll et al., 1998). Calpastatin, which is an endogenous inhibitor (Ca2+ dependent cysteine proteinase), plays a central role in regulation of calpain activity in cells (Forsberg et al., 1989) and is considered one of the major modulators of the calpain. Therefore, calpastatin may affect proteolysis of myofibrils due to regulation of calpain, which can initiate postmortem degradation of myofibril proteins (Goll et al., 1992; Huff-Lonergan et al., 1996). At the protein structural level, calpastatin is a five-domain inhibitory protein (Killefer and Koohmaraie, 1994). Of the five domains, the N-terminal Leader (L) domain does not appear to have any calpains inhibitory activity, but may be involved in targeting or intracellular localization (Takano et al., 1999), while the other domains (I-IV) are highly homologous and are each independently capable of inhibiting calpains (Cong et al., 1998). This indicates that the inhibitory domains of calpastatin contain three highly conserved regions, A, B and C, of which A and C, bind calpain in a strictly Ca2+-dependent manner but have no inhibitory activity, whereas region B inhibits calpain on its own. It was also found that the removal of the XL domain played a regulatory role by altering phosphorylation patterns on the protein (Takano et al., 1999). These observations suggested that genes coding for calpain and calpastatin may be considered as candidate genes in muscle growth efficiency and meat quality in sheep. Zandi fat-tailed sheep are adapted to the dry and harsh climatic conditions, and are primarily raised for mutton, with milk and wool being of secondary importance (Ghafouri-Kesbi and Eskandarinasab, 2008). Given the important roles of the myostatin and calpain-calpastatin system in meat quality, it is of great interest to study the genes encoding the biohemical pathways related to these proteins. Because there has been no selection program to improve meat quality in Zandi sheep breed, it was hypothesied that the genes encoding the myostatin and calpastatin proteins may show high diversity in Zandi sheep population. Therefore, the aim of the present study was to analyze the polymorphism of the MSTN and CAST genes in Zandi sheep breed.
Materials and methods
Animals and Sampling
Random blood samples were collected from 100 Zandi sheep from three populations involving: the Zandi Sheep Breeding Station, situated in Khojir National Park and Saveh and Damavand cities of Iran (Figure 1). Approximately, a 3-mL blood sample was collected from the jugular vein in EDTA-containing tube and stored at -20°C.
DNA extraction and PCR amplification
Genomic DNA was isolated by using DNA extraction kit (Diatom, GenFanAvaran, Iran) which was based on Boom et al. (1990) method. The quantity of DNA was determined by measuring the absorbance at 260 nm and the concentration, purity and quality were determined by measuring the absorbance at 260/280 nm and 230/260 ratios using a NanoDropTM 1000 spectrophotometer (Thermo Scientific, USA). The DNA extractions were appropriately labeled and stored at -20°C for analysis. Two loci were selected for this study. The exon 3 from the ovine myostatin gene was amplified to produce a 337 bp product using primers based on the sequence of the ovine myostatin genes (Table 1). The polymerase chain reaction (PCR) was performed using a buffer PCR 1X, 200 μM dNTPs, 1μM MgCl2, 0.4 pmol of each primer, 0.7 U Taq DNA polymerase, 100 ng ovine gnomic DNA and H2O up to a total volume of 20 μL. Thirty-five cycles of preliminary denaturation at 95°C (5 min), denaturation at 94°C (30 sec), annealing at 62°C (40 sec), extension at 72°C (40 sec) and final extension at 72°C (5 min). The PCR products were separated by 1% (w/v) agarose gel electrophoresis. The exon and intron regions from a portion of the first repetitive domain of the ovine calpastatin gene were amplified to produce a 622 bp fragment using primers based on the sequence of the bovine and ovine calpastatin genes (Table 1). Polymerase chain reaction was performed using a buffer PCR 1X, 200 μM dNTPs, 1.5 μM MgCL2, 0.4 pmol of each primer, 1 U Taq DNA polymerase, 50 ng ovine gnomic DNA and H2O up to a total volume of 20 μL.
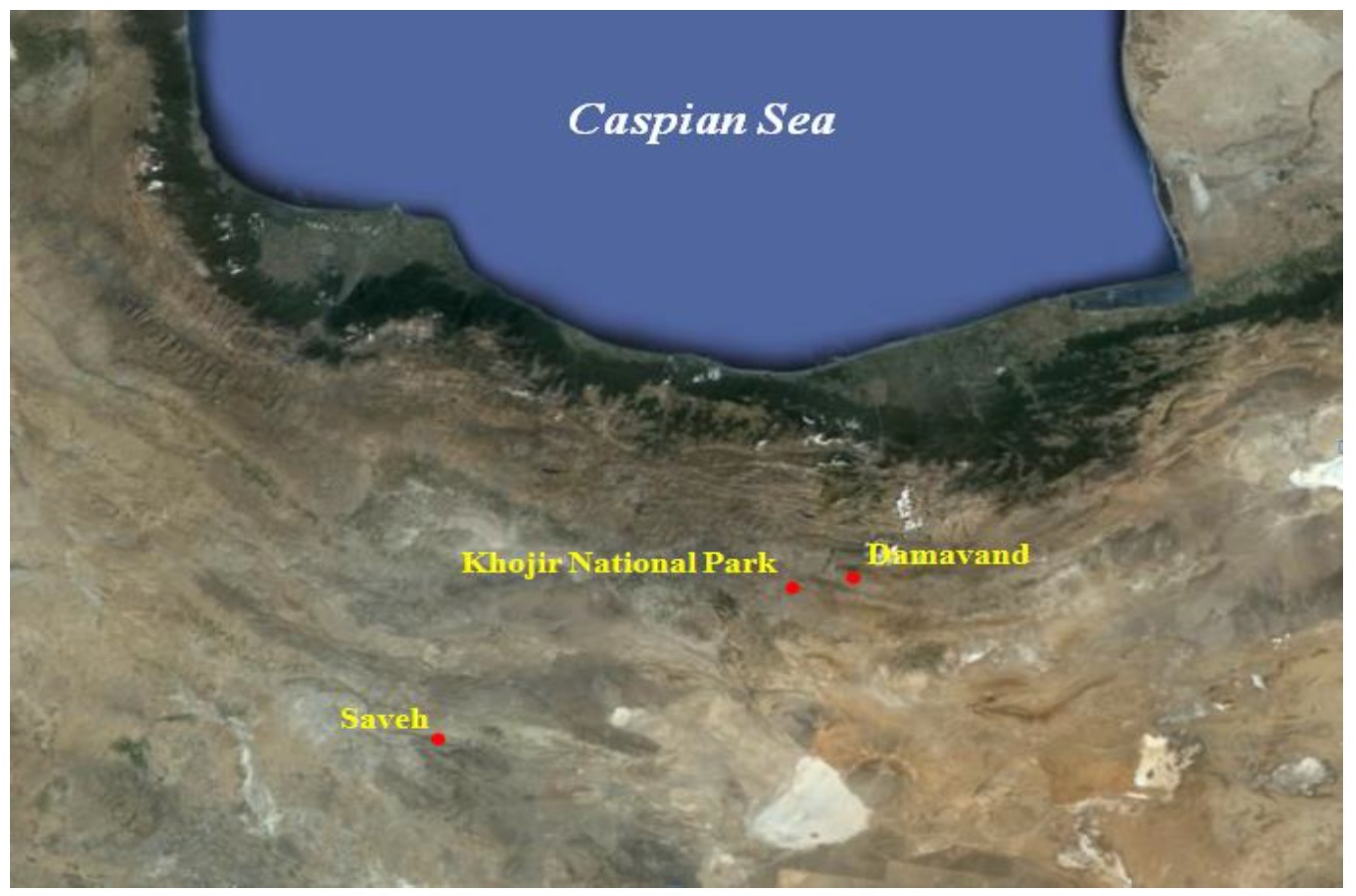
Figure 1. Geographical location of the Zandi sheep populations studied
Table 1. Primers used for MSTN and CAST genes amplification (suggested by Smith et al., 1997; Palmer et al., 1999)
|
PCR product size (bp) |
Locus Primer sequence |
|
337 |
MSTN |
|
|
F: 5'- CCGGAGAGACTTTGGGCTTGA |
|
|
R: 5'-TCATGAGCACCCACAGCGGTC |
|
622 |
CAST |
|
|
F: 5'-TGGGGCCCAATGACGCCATCGATG
|
|
|
R: 5'-GGTGGAGCAGCACTTCTGATCACC |
Thirty-five cycles of preliminary denaturation at 95°C (5 min), denaturation at 94°C (30 sec), annealing at 62°C (40 sec), extension at 72°C (50 sec) and final extension at 72°C (5 min). The PCR products were separated by 1% (w/v) agarose gel electrophoresis.
Enzyme digestion and statistical analysis
The amplified fragment of MSTN was digested with HhaIII enzyme. Digestion was conducted at 37°C for 12-16 h and in a 20 μL reaction solution including 12.5 μL distilled H2O, 2 μL buffer10X, 0.5 μL (5 units) restriction endonucleases (HhaIII) and 5 μL PCR product solutions. The digestion products were electrophoresed on 1.5% agarose gel in 1X TBE and visualized by GelRed staining for 45 min at 80V. The amplified fragment of CAST was digested with MspI enzyme. Digestion was conducted at 37°C for 12-16 h and in a 20 μL reaction solution including 12.5 μL distilled H2O, 2 μL buffer10X, 0.5 μL (5 units) restriction endonucleases (MspI) and 5 μL PCR product solutions. The digestion products were electrophoresed on 1.5% agarose gel in 1X TBE and visualized by GelRed staining for 1 h at 80V. The genotypic and allelic frequencies and other population genetic analyses were conducted using the PopGene software (Yeh et al., 1999).
Results
A 337 bp fragment from exon 3 of MSTN locus was amplied (Figure 2). Thee HaeIII restriction enzyme was used to digest the PCR products. The HaeIII digests the m allele, but not M allele. Digestion of the m allele produced three fragments of 83, 123 and 131 bp and all samples showed the mm genotype (Figure 3). As a result, all of them were monomorphic. The amplified CAST gene resulted in a DNA fragment with 622 bp including the sequences of exon and intron regions from a portion with PCR technique (Figure 4). Two alleles (A and B) were observed, resulting in three genotypes. The MspI digestion of the PCR products produced digestion fragments of 306 bp and 259 bp. The animals with both alleles were assigned as AB genotype, whereas those possessing only A or B alleles as AA or BB genotypes, respectively. The AA genotype showed the two- band pattern (bands of approximately 306 and 259 bp). The AA genotype showed one band pattern (approximately 565 bp), while AB animals displayed a pattern with all three bands (565, 306, 259) (Figure 5). The genotypes of all animals were used to determine the allele frequencies. The A and B allele frequencies were 0.78 and 0.22, respectively. The observed genotype frequencies were 0.60 for AA, 0.04 for BB and 0.36 for AB (Table 2). This sheep population was in Hardy-Weinberg equilibrium for the CAST gene. The observed and expected heterozygosity were 0.360 and 0.344, respectively. The effective allele and true allele estimates were 1.522 and 2.000, respectively (Table 3). This difference between effective and observed allele number and low diversity is due to more frequency of A allele compared to B allele, that reduced frequency in any locus.
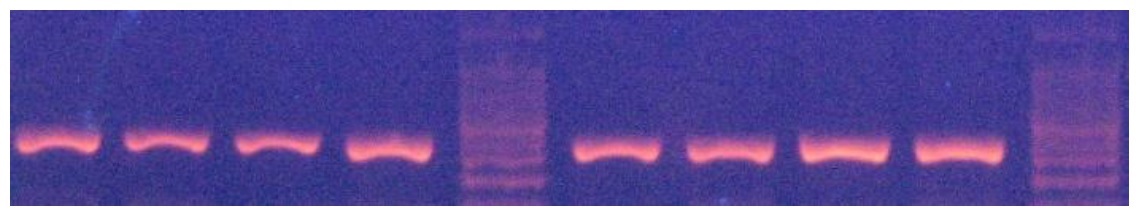
Figure 2. PCR products of MSTN gene in Zandi sheep (Size obtained: 337 bp)
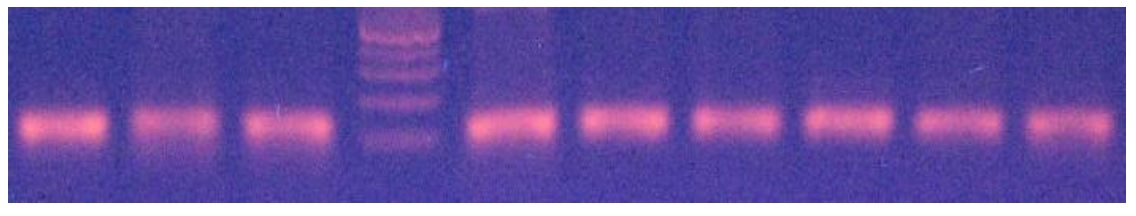
Figure 3. MSTN genotyping in Zandi sheep by PCR-RFLP method (2 % agaros gel)
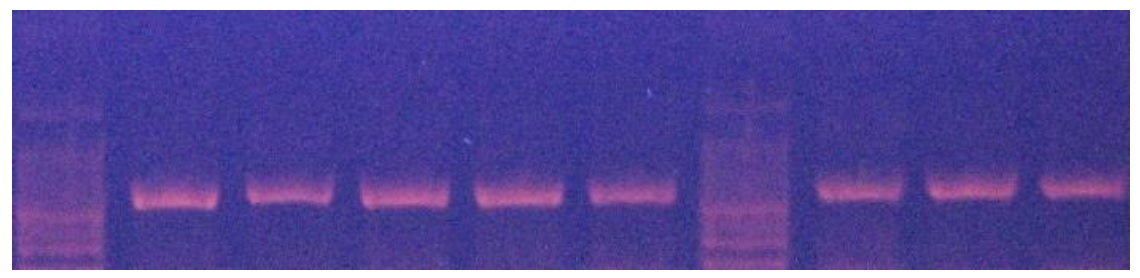
Figure 4. PCR products of CAST gene in Zandi sheep (Size obtained: 622 bp)
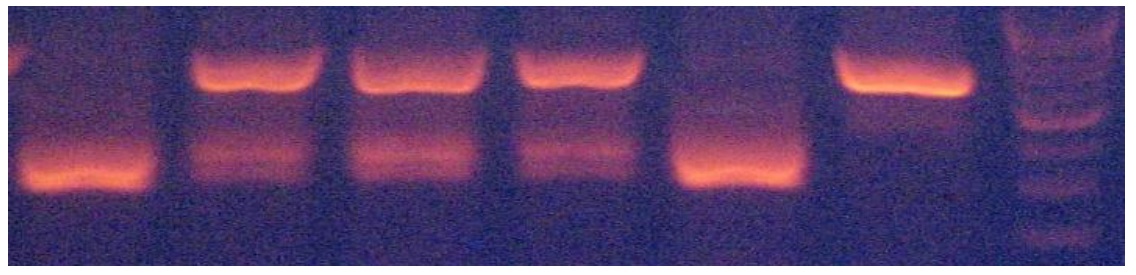
Figure 5. CAST genotyping by PBR method in Zandi sheep (1.5% agaros gel)
Table 2. Genotypic and allelic frequencies of CAST locus in Zandi sheep
|
Allelic frequencies |
Genotypic frequencies |
|||
|
B |
A |
BB |
AB |
AA |
|
0.22 |
0.78 |
0.04 |
0.36 |
0.60 |
Table 3. The observed number of alleles (Na), effective number of alleles (Ne), and observed and expected heterozygosity in CAST locus
|
Heterozygosity |
Homozygosity |
Ne |
Na |
|
0.344 |
0.360 |
1.522 |
2.000 |
Discussion
Candidate genes are known to have biological functions related to the development or physiology of an important trait. We observed no variability in MSTN locus. Similar to our findings, this locus did not show polymorphism in Dalagh sheep (Ahani Azari et al., 2012). On the contrary, Soufy et al. (2009) observed polymorphism for MSTN gene in Sanjabi sheep. This inconsistency may be ascribed to breed differences, population and sample size, mating strategies, geographical position effect and frequency distribution of genetic variants. Although myostatin locus was monomorphic in this population, results showed acceptable polymorphism for calpastatin and calpain loci, which may open interesting prospects for future selection programs, especially using marker-assisted selection for improving weight gain and meat quality. Moreover, This locus proved to have high polymorphism in Lori, Arabi, Dalagh, Zel and other native breeds in Iran (Mohammadi et al., 2008). In the present study, two alleles (A and B) and three genotypes (AA, AB and BB) were observed for CAST gene. Variation in non-coding and coding regions of the ovine CAST gene has been reported by several researchers (Roberts et al., 1996; Palmer et al., 1998; Palmer et al., 2000; Zhou et al., 2007). Study of polymorphism on the same region of the CAST gene in Kurdi sheep by PCR-SSCP revealed three genotypes including aa, ab and ac (Nassiry et al., 2006). The polymorphism in the exon 1 of the CAST in sheep was also reported by other researchers using PCR-RFLP technique (Palmer et al., 1998; Mohammadi et al., 2008; Gabor et al., 2009). In goats and cattle, the exon 6 of CAST gene was investigated for polymorphism and a number of allelic variants were identified in these species (Zhou et al., 2007; Zhou and Hickford 2008). Higher frequencies of CAST gene A allele compared to the B allele have been reported in Nellore (0.66), Rubia Gallega (0.72), Canchim (0.62), Brangus (0.78) and Pardo Suico (0.80) cattle (Asadi and Khederzadeh, 2015). There are several studies on the association of CAST gene polymorphism and meat quality in animals. Schenkel et al. (2006) reported a significant association between C allele of bovine CAST gene and meat tenderness. Kuryl et al. (2003) reported that CAST gene may be considered as a candidate gene for pig carcass quality. Association between the D and F alleles of porcine CAST gene and meat quality traits was also reported by Kapelanski et al. (2004). Palmer et al. (1999) found allelic frequencies of 0.69 and 0.70 for A allele in Dorset Down and Coopworth, respectively, which was in close agreement with the frequency of the A allele in Makoei sheep in the present study. In contrast, they reported that frequencies of A and B alleles in Corriedale and Ruakura were 0.27 and 0.41, respectively. Different frequencies for the alleles of the CAST gene have been reported in Iranian Baluchi sheep with 0.70 for A allele, 0.08 for B allele, and 0.22 for C allele. The BC and CC genotypes, which presented, respectively, 0.03 and 0.04 frequencies in Baluchi sheep (Tahmoorespur et al., 2007), were not observed in Makoei sheep. Two allelic systems of polymorphic variants (M and N) in the region of ovine CAST locus were described by PCR-RFLP method (Palmer et al., 1998; Shahroodi et al., 2005). According to Palmer et al. (1998), allelic frequencies were 77% and 12% for the M and N in Corriedale sheep, respectively. Therefore, CAST may be a suitable for gene assistant selection.
Conclusions
The Zandi sheep breed showed no genetic diversity for the MSTN gene, but the polymorphism found in the CAST gene may be helpful in selection programs for genetic improvement of meat traits. However, before application in the genetic improvement of the indigenous sheep breeds, the association of these polymorphisms with meat traits need to be established.
Acknowledgment
The study was supported by the Department of Genetics and Biotechnology of the Islamic Azad University, Varamin-Pishva Branch, Iran.